NGS is a high-throughput technology capable of sequencing genomic strands in parallel7
- Allows multiplexing, whereby it can simultaneously interrogate multiple potentially actionable targets in a single sample7,8
- Detects fusions among any of the 3 NTRK genes (NTRK1, NTRK2, or NTRK3 ) and a gene partner (known or novel), regardless of breakpoint or fusion partner, depending on library prep method2
- DNA- and RNA-based NGS methods exist; however, RNA-based methods are preferred, as they avoid the difficulties of sequencing large intronic regions in NTRK gene fusions2
IHC is a technique of visualizing proteins in histological tissue sections
RT-PCR is targeted amplification of a predefined region of interest on an RNA template. Products are visualized using numerous downstream techniques, including gel electrophoresis or real-time quantitative PCR, that can be used to monitor the response of cancer cells to drugs16:
PLANNING FOR
YOUR PRACTICE
Several methods exist to detect NTRK gene fusions. RNA-based NGS methods may be ideal because they can identify the many known and novel fusion partners and variable structural properties of NTRK gene fusions8
DNA, deoxyribonucleic acid; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; NGS, next-generation sequencing; NTRK, neurotrophic tyrosine receptor kinase; RNA, ribonucleic acid; RT-PCR, reverse-transcription polymerase chain reaction; TRK, tropomyosin receptor kinase.
References
- Vaishnavi A, Le AT, Doebele RC. Cancer Discov. 2015;5(1):25-34. Return to content
- Schram AM, Chang MT, Jonsson P, Drilon A. Nat Rev Clin Oncol. 2017;14(12):735-748. Return to content
- Wilson KD, Schrijver I. Transitioning diagnostic molecular pathology to the genomic era: cancer somatic mutation panel testing. In: Yousef GM, Jothy S, eds. Molecular Testing in Cancer. New York, NY: Springer Science+Business Media, LLC; 2014:3-11. Return to content
- O’Connor C. Fluorescence in situ hybridization (FISH). Nat Educ. 2008;1(1):171. Return to content
- Teixidó C, Giménez-Capitán A, Molina-Vila MÁ, et al. RNA analysis as a tool to determine clinically relevant gene fusions and splice variants. Arch Pathol Lab Med. 2018;142:474-479. Return to content
- Kroneis T, Jonasson E, Andersson D, Dolatabadi S, Ståhlberg A. Global preamplification simplifies targeted mRNA quantification. Sci Rep. 2017;7:45219. Return to content
- Serrati S, De Summa S, Pilato B, et al. Next-generation sequencing: advances and applications in cancer diagnosis. Onco Targets Ther. 2016;9:7355-7365. Return to content
- Kummar S, Lassen UN. Target Oncol. 2018;13(5):545-555. Return to content
- Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19(3):341-365. Return to content
- Basho RK, Eterovic AK, Meric-Bernstam F. Clinical applications and limitations of next-generation sequencing. Am J Hematol Oncol. 2015;11(3):17-22. Return to content
- Vendrell JA, Taviaux S, Béganton B, et al. Detection of known and novel ALK fusion transcripts in lung cancer patients using next-generation sequencing approaches. Sci Rep. 2017;7:1-11. Return to content
- Fitzgibbons PL, Cooper K. Immunohistochemistry of biomarkers. In: Cagle PT, Allen TC, eds. Basic Concepts of Molecular Pathology. Molecular Pathology Library Series. New York, NY: Springer Science+Business Media, LLC; 2009:133-137. Return to content
- Church AJ, Calicchio ML, Nardi V, et al. Recurrent EML4-NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod Pathol. 2018;31(3):463-473. Return to content
- Hechtman JF, Benayed R, Hyman DM, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol. 2017;41(11):1547-1551. Return to content
- Halling KC, Wendel AJ. In situ hybridization; principles and applications. In: Cagle PT, Allen TC, eds. Basic Concepts of Molecular Pathology. Molecular Pathology Library Series. New York, NY: Springer Science+Business Media, LLC; 2009:109-118. Return to content
- Olsen JL. Polymerase chain reaction. In: Vohr H-W, ed. Encyclopedia of Immunotoxicology. Berlin, Germany: Springer-Verlag; 2016:715-720. Return to content
- Argani P, Fritsch M, Kadkol SS, Schuster A, Beckwith JB, Perlman EJ. Detection of the ETV6-NTRK3 chimeric RNA of infantile fibrosarcoma/cellular congenital mesoblastic nephroma in paraffin-embedded tissue: application to challenging pediatric renal stromal tumors. Mod Pathol. 2000;13(1):29-36. Return to content
An algorithmic approach for NTRK gene fusion detection
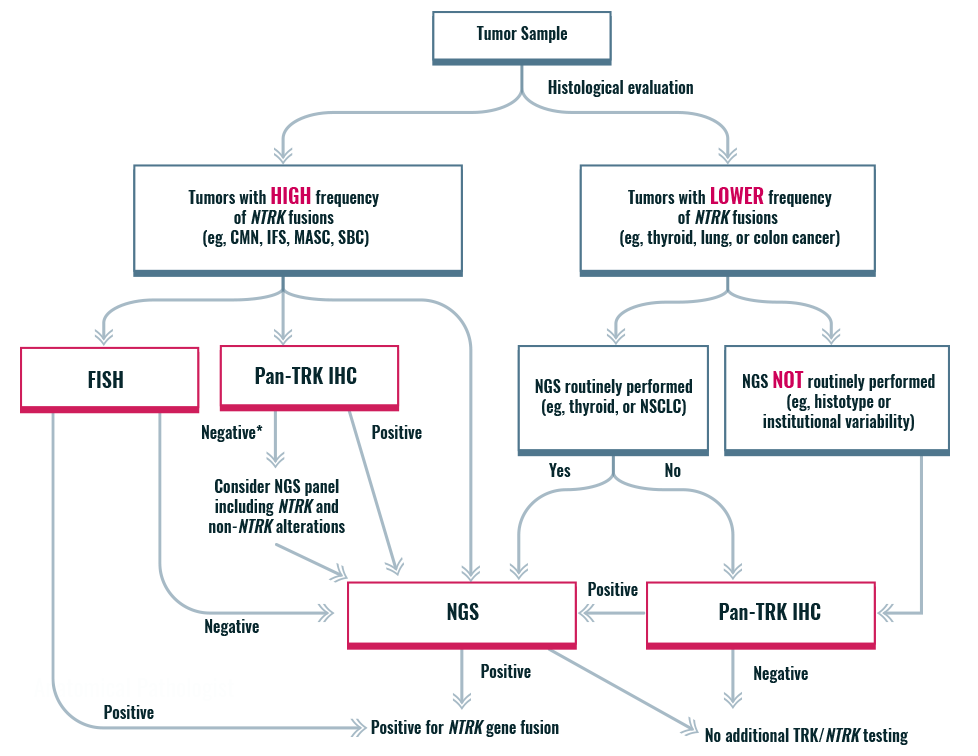
*If histology typical, then confirmation by NGS recommended
CMN, congenital mesoblastic nephroma; FISH, fluorescence in situ hybridization; IFS, infantile fibrosarcoma; IHC, immunohistochemistry; MASC, mammary analogue secretory carcinoma; NGS, next-generation sequencing; NSCLC, non-small cell lung cancer; NTRK, neurotrophic tyrosine receptor kinase; SBC, secretory breast cancer; TRK, tropomyosin receptor kinase.
References
- Penault-Llorca F, Rudzinski ER, Sepulveda AR. Testing algorithm for identification of patients with TRK fusion cancer. J Clin Pathol. 2019 May 09. doi: 10.1136/jclinpath-2018-205679 [Epub ahead of print] Return to content
Key considerations for detecting NTRK gene fusions
- RNA-based NGS is preferred to DNA-based NGS, as it enables distinction of in-frame, transcribed gene fusions vs out-of-frame fusions and avoids sequencing large intronic regions associated with NTRK gene fusions1,2
- Not all commercially available NGS panels comprehensively cover NTRK1, NTRK2, and NTRK3 gene fusions3,4
Advantages
PLANNING FOR
YOUR PRACTICE
A variety of commercially available NGS kits can be used to efficiently detect fusions across NTRK1, NTRK2, and NTRK3 genes. Sample requirements (DNA vs RNA) and bioinformatic requirements vary among these kits and associated enrichment methods with these kits impact each kit’s detection capabilities11
- Nucleic acid extraction (from various sample types)
- Simultaneous amplification of regions of interest (hybrid capture– vs amplicon-based target enrichment)
- Massively parallel sequencing (instrument reads massive amounts of DNA fragments simultaneously)
- Sequence analysis (complex bioinformatic analysis)
- Computer software aligns, filters noise, and compares sequence to reference genome
DNA, deoxyribonucleic acid; NGS, next-generation sequencing; NTRK, neurotrophic tyrosine receptor kinase; RNA, ribonucleic acid; TRK, tropomyosin receptor kinase.
References
- Serrati S, De Summa S, Pilato B, et al. Next-generation sequencing: advances and applications in cancer diagnosis. Onco Targets Ther. 2016;9:7355-7365. Return to content
- Schram AM, Chang MT, Jonsson P, Drilon A. Nat Rev Clin Oncol. 2017;14(12):735-748. Return to content
- QIAGEN. GeneRead™ QIAact Lung All-in-One UMI Assay [product profile]; 2017. https://amp.qiagen.com/2017/wp-content/uploads/sites/2/2017/11/PROM-11408-001_1108382_PP_GeneRead_QIAact_LungAiO_0917_WW.pdf. Accessed March 3, 2019. Return to content
- QIAGEN. GeneRead™ QIAact Lung RNA Fusion UMI Panel Handbook. April 2018. https://QIAGEN.+GeneRead%E2%84%A2+QIAact+Lung+RNA+Fusion+UMI+Panel+Handbook&oq=QIAGEN.+GeneRead%E2%84%A2+QIAact+Lung+RNA+Fusion+UMI+Panel+Handbook&aqs=chrome..69i57.2540j0j4&sourceid=chrome&ie=UTF-8. Accessed March 3, 2019. Return to content
- Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing–based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19(3):341-365. Return to content
- Abel H, Pfeifer J, Duncavage E. Translocation detection using next-generation sequencing. In: Kulkarni S, Pfeifer J, eds. Clinical Genomics. New York, NY: Elsevier/Academic Press; 2015:151-164. Return to content
- Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4-23. Return to content
- Teixidó C, Giménez-Capitán MA, Molina-Vila MÁ, et al. RNA analysis as a tool to determine clinically relevant gene fusions and splice variants. Arch Pathol Lab Med. 2018;142:474-479. Return to content
- Wilson KD, Schrijver I. Transitioning diagnostic molecular pathology to the genomic era: cancer somatic mutation panel testing. In: Yousef GM, Jothy S, eds. Molecular Testing in Cancer. New York, NY: Springer Science+Business Media, LLC; 2014:3-11. Return to content
- Hechtman JF, Benayed R, Hyman DM, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol. 2017;41(11):1547-1551. Return to content
- Kummar S, Lassen UN. Target Oncol. 2018;13(5):545-556. Return to content
- Gagan J, Van Allen EM. Next-generation sequencing to guide cancer therapy. Genome Med. 2015;7(1):80. Return to content
Key considerations for detecting TRK fusion proteins
- Pan-TRK IHC can detect all 3 fusion proteins–TRKA,TRKB, and TRKC–in addition to wild-type TRK proteins1
- Strong positive staining for TRK protein is not always due to the presence of an NTRK gene fusion and can have a negative impact on the specificity of the assay performance2
- Some cancers are associated with high physiological expression of wild-type TRK protein, and are not the result of a gene fusion, thus limiting the usefulness of IHC methodology2,3
Challenges
- Typically limited to investigating only one analyte at a time in the clinical setting5
- Requires experienced and qualified pathologists to correctly evaluate the extent or degree of intensity of the staining to determine whether the test is positive2
- Lack of standardization is common in preanalytical, analytical, and postanalytical aspects of testing2
- Low specificity for TRK fusion proteins3
PLANNING FOR
YOUR PRACTICE
Because IHC measures protein expression, results should be confirmed using NGS to ensure abnormal expression is associated with a fusion protein vs wild-type TRK protein2
TESTING WORKFLOW OVERVIEW6
- Tissue fixed and embedded within paraffin block
- Only 10% NBF is recommended
- Fix immediately; delayed time to fixation may result in tissue degradation
- Sections cut and placed on a microscope slide
- Cut sections approximately 3-6 μm in thickness
- Mount on positively charged slides
- Sections labeled with antibody
- Sections stained and visualized
- Immediate staining is recommended, as antigenicity may diminish over time
- Consult assay for instruction for recommended staining protocol
Using pan-TRK IHC as a screening tool
Pan-TRK IHC detects TRKA, TRKB, and TRKC, but lacks specificity to serve as stand-alone test6
- IHC detects both wild-type and fusion proteins, thus protein expression may not be directly correlated to the presence of an NTRK gene fusion
- After a positive TRK IHC test, confirm NTRK gene fusions via a validated molecular test (eg, NGS)
Commercially available assays and antibodies can detect TRK proteins: TRKA, TRKB, TRKC
MANUFACTURER: Roche Tissue Diagnostics/Ventana
ANTIBODY/ASSAY
VENTANA pan-TRK (EPR17341) Assay
CLASSIFICATION/SPECIFIC
IVD/CE-IVD
Assay detects C-terminal region of TRKA, TRKB, TRKC6
MANUFACTURER: Abcam
ANTIBODY/ASSAY
Anti-TRKA+ TRKB+TRKC [EPR18413]
CLASSIFICATION/SPECIFIC
Research use only
Synthetic peptide within Human TRKC amino acid to the C-terminus7
MANUFACTURER: Abcam
ANTIBODY/ASSAY
Anti-TRKA+ TRKB+TRKC [EPR17341]
CLASSIFICATION/SPECIFIC
Research use only
Synthetic peptide within Human TRKC amino acid to the C-terminus8
MANUFACTURER: Cell Signaling Technology
ANTIBODY/ASSAY
TRK (pan) (A7H6R) Rabbit mAb
CLASSIFICATION/SPECIFIC
Research use only
Rabbit mAb detects endogenous levels of TRKA, TRKB, and TRKC9
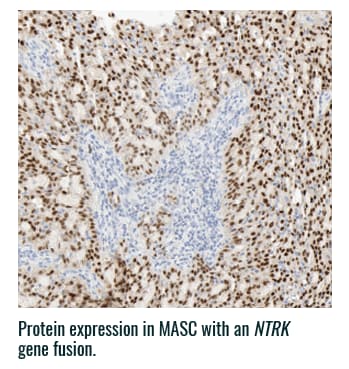
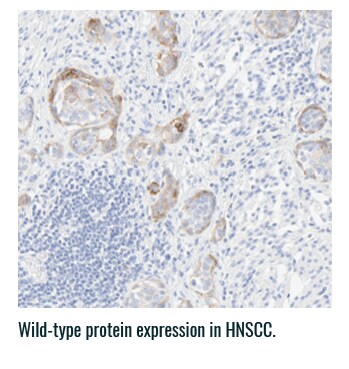
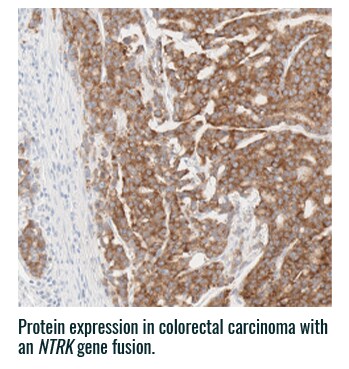
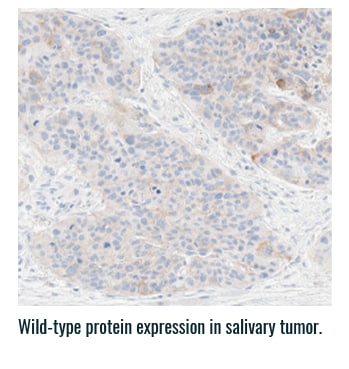
Pan-TRK staining varies in intensity and percentage across tumor types3
- Pan-TRK IHC may be helpful in tumor types with low prevalence of wild-type expression (such as CRC and papillary thyroid) where it can serve as a screening tool to identify NTRK gene fusion-harboring tumors3,6
- However, for tumors with high prevalence of wild-type TRK protein expression, IHC is less helpful, as staining intensity and percentage for fusion proteins cannot be distinguished from wild-type expression6
PAN-TRK STAINING ACROSS TUMOR TYPES10,*
Ventana Pan-TRK [EPR17341] Assay.
HNSCC, head and neck squamous cell carcinoma; IHC, immunohistochemistry; MASC, mammary analogue secretory carcinoma; NBF, neutral buffered formalin; NGS, next-generation sequencing; NSCLC, non-small cell lung cancer; NTRK, neurotrophic tyrosine receptor kinase; TRK, tropomyosin receptor kinase.
References
- Hechtman JF, Benayed R, Hyman DM, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol. 2017;41(11):1547-1551. Return to content
- Fitzgibbons PL, Cooper K. Immunohistochemistry of biomarkers. In: Cagle PT, Allen TC, eds. Basic Concepts of Molecular Pathology. Molecular Pathology Library Series. New York, NY: Springer Science+Business Media, LLC; 2009:133-137. Return to content
- Feng J, Ebata K, Hansen F, et al. TRK wild-type and fusion protein expression in solid tumors: characterization by immunohistochemistry and in situ hybridization. Poster presented at: MAP 2018—Molecular Analysis for Personalised Therapy. European Society for Medical Oncology Congress (ESMO); September 15, 2018; Paris, France. Return to content
- Wilson KD, Schrijver I. Transitioning diagnostic molecular pathology to the genomic era: cancer somatic mutation panel testing. In: Yousef GM, Jothy S, eds. Molecular Testing in Cancer. New York, NY: Springer Science+Business Media, LLC; 2014:3-11. Return to content
- Halling KC, Wendel AJ. In situ hybridization: principles and applications. In: Cagle PT, Allen TC, eds. Basic Concepts of Molecular Pathology. Molecular Pathology Library Series. New York, NY: Springer Science+Business Media, LLC;2009:118-127. Return to content
- VENTANA pan-TRK (EPR17341) Assay [product information]. Mannheim, Germany: Roche Diagnostics GmbH: 2018. Return to content
- Abcam [product data sheet]. Anti-TrkA+TrkB+TrkC antibody [EPR18413] ab189903. https://www.abcam.com/pan-trk-antibody-epr18413-ab189903. Accessed February 27, 2019. Return to content
- Abcam [product data sheet]. Anti-TrkA+TrkB+TrkC antibody [EPR17341] ab181560. https://www.abcam.com/pan-trk-antibody-epr17341-ab181560.html. Accessed February 27, 2019. Return to content
- Cell Signaling Technology [product data sheet]. Trk (pan) (A7H6R) Rabbit mAb. https://media.cellsignal.com/pdf/92991.pdf. Accessed February 27, 2019. Return to content
- Ventana Medical System, Inc. VENTANA pan-TRK (EPR17341) Assay: your assay choice matters [brochure]. Tucson, AZ: 2018. Return to content
Key considerations for detecting NTRK gene fusions
- Does not establish if rearrangement results in an oncogenic fusion1
- Diverse range of potential NTRK gene fusion partners associated with the 3 genes (NTRK1, NTRK2, and NTRK3 ) will make FISH a labor-intensive and inefficient option1
- Limited biopsy material may present a barrier to detecting all relevant NTRK gene fusions if looking for the novel fusion partner2
Challenges
PLANNING FOR
YOUR PRACTICE
FISH does not establish if a rearrangement results in a functional fusion, thus results should be interpreted with caution. Due to the range of potential NTRK gene fusion partners, this will become highly labor intensive, as individual reactions for each NTRK gene will be required1
TESTING WORKFLOW OVERVIEW2
- Cells fixed
- Cells placed on a microscope slide
- Break-apart/dual-colored probe hybridized to the cells (overnight incubation)
- Analysis using fluorescence microscope
FISH, fluorescence in situ hybridization; NTRK, neutrophic tyrosine receptor kinase.
References
- Church AJ, Calicchio ML, Nardi V, et al. Recurrent EML4-NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod Pathol. 2018;31(3):463-473. Return to content
- Halling KC, Wendel AJ. In situ hybridization: principles and applications. In: Cagle PT, Allen TC, eds. Basic Concepts of Molecular Pathology. Molecular Pathology Library Series. New York, NY: Springer Science+Business Media, LLC; 2009:109-118. Return to content
- Wilson KD, Schrijver I. Transitioning diagnostic molecular pathology to the genomic era: cancer somatic mutation panel testing. In: Yousef GM, Jothy S, eds. Molecular Testing in Cancer. New York, NY: Springer Science+Business Media, LLC; 2014:3-11. Return to content
- Cheng L, Zhang S, Wang L, MacLennan GT, Davidson DD. Fluorescence in situ hybridization in surgical pathology: principles and applications. J Path Clin Res. 2017;3(2):73-99. Return to content
Key considerations for detecting NTRK gene fusions
PLANNING FOR
YOUR PRACTICE
RT-PCR is already used to detect specific fusion combinations associated with certain cancer indications; however, it cannot be used for the detection of novel fusion partners1
TESTING WORKFLOW OVERVIEW4,6
- RNA extraction and ctDNA synthesis (from various sample types and platforms)
- RT-PCR reaction (counts normal and mutant copies amplified in real time, ie, as the PCR reaction takes place)
ctDNA, chloroplast deoxyribonucleic acid; NTRK, neurotrophic tyrosine receptor kinase; PCR, polymerase chain reaction; RNA, ribonucleic acid; RT-PCR, reverse-transcription polymerase chain reaction.
References
- Church AJ, Calicchio ML, Nardi V, et al. Recurrent EML4-NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod Pathol. 2018;31(3):463-473. Return to content
- Xu T, Wang, H, Huang X, et al. Gene fusion in malignant glioma: an emerging target for next-generation personalized treatment. Transl Oncol. 2018;11(3):609-618. Return to content
- Vaishnavi A, Le AT, Doebele RC. Cancer Discov. 2015;5:25-34. Return to content
- Olsen JL. Polymerase chain reaction. In: Vohr H-W, ed. Encyclopedia of Immunotoxicology. Berlin, Germany: Springer-Verlag; 2016:715-720. Return to content
- Argani P, Fritsch M, Kadkol SS, Schuster A, Beckwith JB, Perlman EJ. Detection of the ETV6-NTRK3 chimeric RNA of infantile fibrosarcoma/cellular congenital mesoblastic nephroma in paraffin-embedded tissue: application to challenging pediatric renal stromal tumors. Mod Pathol. 2000;13(1):29-36. Return to content
- Peter M, Gilbert E, Delattre O. A multiplex real-time PCR assay for the detection of gene fusions observed in solid tumors. Lab Invest. 2001;81:905-912. Return to content